Improving standard of Prostate Cancer Diagnosis and Care while increasing MRI revenues
May 01, 2024
Health IT
MRI
Ultrasound
Sponsored
Industry Excellence Profile

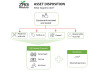
Standalone Performance AUROC curve of Bot Image's ProstatID AI Software obtained during clinical studies using retrospective Imaging and Pathology data from various MRI vendors and locations
Improving standard of Prostate Cancer Diagnosis and Care while increasing MRI revenues and providing Urologists with Targeting roadmap.
The standard of care (SOC) of prostate cancer (PCa) diagnosis and treatment is out of date. MRI has long ago proven to have the sensitivity and specificity to detect most prostate cancers at an early stage. Furthermore, studies published as early as RSNA 2017 have indicated that non-contrast MRI (bi-parametric MRI) has proven useful for fast, low-cost, detection of PCa with nearly identical outcomes as contrast (mpMRI) [1]. This would imply that bpMRI would meet the AMA criteria for screening because it is easy to perform and interpret, acceptable, accurate, reliable sensitive and specific; and further meet Wilson’s criteria for screening in that it must also be readily available and relatively inexpensive; which bpMRI is.
However; today, prostate MRI is Not Easy to interpret, nor is bpMRI widely accepted. PCa MRI is poorly interpreted as a whole within the industry. Urologists have increasingly become disenchanted with the use of MRI in general because of the many false positives and missed cancers that they detect after receiving an MRI report. Urologists typically start with a standard 12-core biopsy and supplement it with additional sampling of radiographic “hot spots” using cognitive fusion.
Another reason for the SOC being out of date is because it is established via the NCCN (Vers. 2, 2024, 03/06/2024) Guidelines for Prostate Cancer Early Detection, and guidelines such as these typically lag behind the scientific and technical advancements by many years. As testament to that, consider that throughout the entire process, the NCCN relies exclusively upon PSA tests and DREs for early detection evaluation despite the facts that neither, nor their combination, has a sensitivity-specificity significantly greater than 50%.
The guidance states further; after repeat PSAs, DREs, then consider mpMRI or biomarkers that “improve the specificity of screening” such as Select MDx, 4Kscore, ExoDx, etc. prostate tests.
PAUSE: Regarding these biomarkers - they are very expensive in order to add a risk score to the diagnostic equation. Let’s focus on the benefit. Even if the score is suggestive of prostate cancer, the next step is to get an MRI “if available” and/or proceed with an image-guided biopsy. Here’s the fine print on page 9 of the guidelines. “It is not yet known how such tests could be applied in optimal combination with MRI.”
DISCUSSION:
The argument about a better biomarker has been resolved. MRI plus a proven Detection and Diagnostic AI software can not only provide sensitivity-specificity results in the mid-90th percentile (Standalone AUROC that created physician improvement noted in this reference) [2], but also establish location, size, and classification of the suspicious lesion(s), in addition to the accurate volume of the gland – this without additional data such as the serum PSA or the expensive genetic tests or biomarkers.
The desired results of Prostate Cancer diagnoses are to have more definitive/accurate and early detections and/or confidence that the patient is cancer free. Moving the needle towards attaining these goals is the role of Artificial Intelligence in MRI interpretations.
References:
1) A narrative review of biparametric MRI (bpMRI) implementation on screening, detection, and the overall accuracy for prostate cancer, Greenberg, Koller, Casado, Triche, Krane, Ther Adv Urol, 2022 May 4. Doi 10.1177/17562872221096377.
2) Improving Prostate Cancer Detection with MRI: A Multi-Reader, Multi-Case Study Using Computer-Aided Detection (CAD), Anderson, Mercaldo, Chung, Ulrich, Jones, Harisinghani, Academic Radiology 2022, https://doi.org/10.1016/j.acra.2022.09.009
The standard of care (SOC) of prostate cancer (PCa) diagnosis and treatment is out of date. MRI has long ago proven to have the sensitivity and specificity to detect most prostate cancers at an early stage. Furthermore, studies published as early as RSNA 2017 have indicated that non-contrast MRI (bi-parametric MRI) has proven useful for fast, low-cost, detection of PCa with nearly identical outcomes as contrast (mpMRI) [1]. This would imply that bpMRI would meet the AMA criteria for screening because it is easy to perform and interpret, acceptable, accurate, reliable sensitive and specific; and further meet Wilson’s criteria for screening in that it must also be readily available and relatively inexpensive; which bpMRI is.
However; today, prostate MRI is Not Easy to interpret, nor is bpMRI widely accepted. PCa MRI is poorly interpreted as a whole within the industry. Urologists have increasingly become disenchanted with the use of MRI in general because of the many false positives and missed cancers that they detect after receiving an MRI report. Urologists typically start with a standard 12-core biopsy and supplement it with additional sampling of radiographic “hot spots” using cognitive fusion.
Another reason for the SOC being out of date is because it is established via the NCCN (Vers. 2, 2024, 03/06/2024) Guidelines for Prostate Cancer Early Detection, and guidelines such as these typically lag behind the scientific and technical advancements by many years. As testament to that, consider that throughout the entire process, the NCCN relies exclusively upon PSA tests and DREs for early detection evaluation despite the facts that neither, nor their combination, has a sensitivity-specificity significantly greater than 50%.
The guidance states further; after repeat PSAs, DREs, then consider mpMRI or biomarkers that “improve the specificity of screening” such as Select MDx, 4Kscore, ExoDx, etc. prostate tests.
PAUSE: Regarding these biomarkers - they are very expensive in order to add a risk score to the diagnostic equation. Let’s focus on the benefit. Even if the score is suggestive of prostate cancer, the next step is to get an MRI “if available” and/or proceed with an image-guided biopsy. Here’s the fine print on page 9 of the guidelines. “It is not yet known how such tests could be applied in optimal combination with MRI.”
DISCUSSION:
The argument about a better biomarker has been resolved. MRI plus a proven Detection and Diagnostic AI software can not only provide sensitivity-specificity results in the mid-90th percentile (Standalone AUROC that created physician improvement noted in this reference) [2], but also establish location, size, and classification of the suspicious lesion(s), in addition to the accurate volume of the gland – this without additional data such as the serum PSA or the expensive genetic tests or biomarkers.
The desired results of Prostate Cancer diagnoses are to have more definitive/accurate and early detections and/or confidence that the patient is cancer free. Moving the needle towards attaining these goals is the role of Artificial Intelligence in MRI interpretations.
References:
1) A narrative review of biparametric MRI (bpMRI) implementation on screening, detection, and the overall accuracy for prostate cancer, Greenberg, Koller, Casado, Triche, Krane, Ther Adv Urol, 2022 May 4. Doi 10.1177/17562872221096377.
2) Improving Prostate Cancer Detection with MRI: A Multi-Reader, Multi-Case Study Using Computer-Aided Detection (CAD), Anderson, Mercaldo, Chung, Ulrich, Jones, Harisinghani, Academic Radiology 2022, https://doi.org/10.1016/j.acra.2022.09.009
You Must Be Logged In To Post A CommentRegisterRegistration is Free and Easy. Enjoy the benefits of The World's Leading New & Used Medical Equipment Marketplace. Register Now! |
|